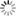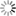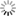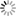INCF Japan Node Session Abstracts
Below are the abstracts of the 10 speakers of this session
1 - STEN GRILLNER
What INCF can do for Neuroscience
International Neuroinformatics Coordinating Facility (INCF) and Nobel Institute for Neurophysiology, Karolinska Institute, Stockholm, Sweden
The goal of the International Neuroinformatics Coordinating Facility, INCF, is to facilitate neuroinformatics research aiming at an understanding of the brain - from the molecular to the cognitive level - and the pathogenic factors underlying the many different diseases of the nervous system, including both psychiatric and neurological aspects [1]. As a consequence, ,INCF is at a cross-road between neuroscience and informatics/physics.
Neuroinformatics provides a very important tool in the analyses of the nervous system. It can be claimed that complex dynamic processes whether at the intracellular or systems level, in which a number of different factors interact, are not understandable without modeling at the single or multi-scale level, nor can the interpretations of such processes and data be tested rigorously without modeling. Similarly, databases with regard to the nervous system are of critical importance for the development and bridging of knowledge in different specific areas, and provide an investigator with easy access to experimental findings in areas which are far outside the investigator’s primary competence. A basic scientist may thus with the help of appropriate data bases relate a finding on for instance an ion channel subtype pathology to a rare clinical condition if the data are accessible – otherwise this connection may take years to discover and the development of therapy will be unnecessarily delayed.
The primary aim of INCF is to develop neuroinformatics infrastructure with regard to modeling, data bases and tool development. To achieve this INCF has developed particular programs in different areas that include multi-scale modeling, digital brain atlasing, standards for metadata and the development of data base friendly hierarchical arranged terminology (ontology). Each program includes the leading international experts in its area, and focuses on scalable, portable and extensible solutions to identified neuroinformatics infrastructure problems and bottle-necks.
The INCF Programs are long-term strategic undertakings. Establishing a program is a multi-step process, which starts with a workshop intended to identify the key issues in the area, and continues into the formation of an oversight committee and task forces. Each program delivers products and services, and develops standards and guidelines for its particular field – through these actions the INCF contributes to the development and maintenance of specific database and other computational infrastructures and support mechanisms. These measures are intended to facilitate the flow of information between researchers in both academia and industry.
The INCF also develops initiatives to coordinate and foster international activities in neuroinformatics, and maintains a web portal to make neuroinformatics resources – research tools and information as well as events, training and jobs – more accessible to the community [2].
By facilitating and strengthening neuroinformatics research, INCF can contribute to a rapid progress in all brain sciences and related fields, and to an understanding of the mechanisms underlying different brain disorders and insights into new potential treatments. Another important role for Neuroinformatics is to form an interface between neuroscience and information technology and robotics – the brain is able to solve many of the problems that man-made technologies still seek to master.
References
1. Bjaalie, J. G., & Grillner, S. (2007), Global Neuroinformatics: The International
Neuroinformatics Coordinating Facility. J Neurosci 27(14), pp 3613- 3615.
2. INCF 2011 – 2015 Strategic Plan (2010), available on www.incf.org, and in hard copy from the secretariat, pp 1-16.
2 - ERIK DE SCHUTTER
New model description standards to facilitate multi-scale modeling
Okinawa Institute of Science and Technology, Japan and University of Antwerp, Belgium
Multi-scale modeling is a tool of critical importance for neuroscience. As computational modeling techniques become integrated with experimental neuroscience, more knowledge can be extracted from existing experimental data. Quantitative models assist in generating experimentally testable hypotheses and in selecting informative experiments. One major challenge in the field is that, because of a wide range of simulation tools being used in the community, it is unlikely that one laboratory can reproduce the results obtained by another group, even if the model is deposited in an openly accessible database. The absence of widely adopted standards for model description also hamper efforts to make existing programs more compatible, reduce opportunities for innovative software development and for benchmarking of existing simulators.
The INCF has started a project to develop a new standard markup language for model description. Based on lessons learned with previous efforts in computational neuroscience and in other fields like systems biology, a concerted effort is made to develop a well-defined but flexible syntax for a self-documenting markup language that will be easy to extend and that can form the basis for specific implementations covering a wide range of modeling scales. The initial effort focuses on describing a growing area of computational neuroscience, spiking networks. This language, called NineML (Network Interchange format for NEuroscience) is based on a layered approach: an abstraction layer allows a full mathematical description of the models, including events and state transitions, while the user layer contains parameter values for specific models. The user layer includes concepts from the four major areas of neuroscience network modeling: neurons, synapses, populations and network connectivity rules. The abstraction layer includes notations for representing hybrid dynamical systems, combining differential equations with event based modeling to describe integrate-and-fire neurons, and abstract mathematical representations of connectivity rules. These abstractions are capable of describing a vast range of network models from the neuroscience literature.
The first official release of NineML is expected by the end of the year. In a next phase NineML will be improved by incorporating community feedback and by expanding the coverage of different spiking network models. Based on this experience new efforts will be launched in the future, using the same language syntax to cover other areas of computational neuroscience, including compartmental models, synaptic microphysiology, cellular mechanics, electrodynamics. Special attention will be given to interoperability issues relevant to multi-scale modeling, where many separate model descriptions may have to be combined and data interfaces need to be defined.
3 - ROBERT W. WILLIAMS
Global Exploratory Analysis of Massive Neuroimaging Collections using Microsoft Live Labs Pivot and Silverlight
Authors: Robert W. Williams, Lei Yan, Xiaodong Zhou, Lu Lu, Arthur Centeno, Leonard Kuan, Michael Hawrylycz, Glenn D. Rosen
Affiliations: Department of Anatomy and Neurobiology, Center for Integrative and Translational Genomics, University of Tennessee Health Science Center, Memphis TN, USA; The Allen Institute for Brain Science, 551 North 34th Street, Seattle, WA, USA; Department of Neurology, Beth Israel Deaconess Medical Center/Harvard Medical School, Boston MA, USA
Introduction: The Mouse Brain Library (www.mbl.org) consists of high resolution images generated for 200 genetically well defined strains of mice, 2200 cases, 8800 Nissl-stained slides, and ~120,000 coronal and horizontal sections of the brain. The MBL includes representatives for most sets of recombinant inbred strains, (BXD, LXS, AXB, BXH, CXB). This collection provides a solid foundation for studies of the genetic control of brain structure, function, and behavior (Lu et al. 2001; Yang et al., 2008; Rosen et al., 2009).
A key challenge is how to deliver massive image collections such as the MBL, Allen Brain Atlas projects, and BrainMaps using modern web services. In this work we tested Microsoft Live Labs Pivot (www.getpivot.com ) as an image distribution portal. Pivot provides a unique method for delivering and exploring very large collections of high-resolution images.
Methods and Results: Large slides of 30-µm celloidin sections were imaged using a 20x objective (NA 0.75) and an Aperio Scanscope CS scanner at an in-plane resolution of 1-µm/pixel and at 150-µm steps along the cutting axis. Images were segmented to extract single sections. SVS format image were converted into Advanced Forensic format (AFF), JPEG2000, and Silverlight deep zoom (DZI) pyramids. A DVI is 2x larger than its JPEG parent. DZIs were assembled into a set of Pivot image collections. Metadata for sorting and display were extracted from MBL and GeneNetwork.org data tables. Metadata types include thousands of genotypes and phenotypes for all strains, as well as data for individual cases (sex, age, body weight, brain weight, litter size). Spatial coordinate tags for each section using INCF Waxholm space and BIRNLex neuroanatomical terms are being added.
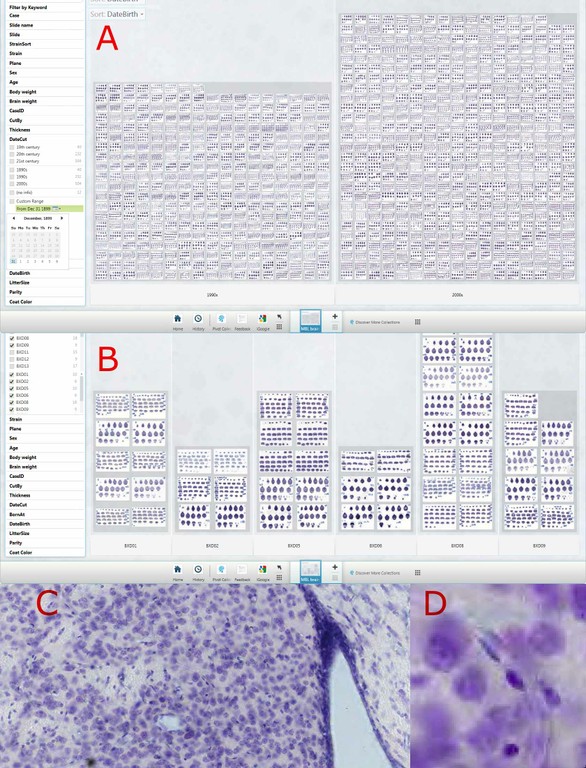
Fig 1. The Pivot interface. A. Forest view of 788 slides in two major categories (1900s, 2000s). B. Tree view of a subset of six strains of mice in which coronal and sagittal sections can be distinguished. C and D are branch and leaf views (progressively higher power zooms) to the level of 1 micron per pixel of a single section (medial striatum).
A Pivot server was installed on a Linux CentOS 5 8-core system (Dell R610) running Tomcat and MySQL. The system was tested on Vista and Windows 7 using a Pivot browser (www.getpivot.com) and runs well on Mac OS X 10.6 using the VMWARE Fusion 3 virtual machine. Link to http://mbl.pivotcollections.org to view the MBL.
The MBL collection is large and unwieldy and our current interface (www.mbl.org) does not provide sufficient sorting flexibility and speed to effectively explore or analyze the collection. A good exploratory interface would provide both forest and tree views and a way to effectively scan the entire collection for variation in traits such as ventricular volume, callosal architecture, cortical lamination, and differences in the cytology in specific nuclei in a matter of minutes, not hours or days. Pivot is optimal for these types of rapid review and exploratory tasks. The collection can be filtered, sorted, and viewed at a wide range of magnifications—from thumbnails of whole slide to full zooms of subnuclei—almost instantly. The collection can be split, filtered, and sorted using a range of continuous and discrete metadata variables (sex, age, strain, genotype, behavior) Limitations with the current Pivot implementation can be divided into two categories: those associated with the interface itself (no nested displays, a limit of 15 displayed categories, no graphic overlay for marking or annotation), and those associated with secondary analytic and statistical functions that would typically be used to test hypotheses (no dynamic output of group statistics nor tests for differences among groups using ANOVA or t tests)
Discussion: Pivot is a superb web service architecture that provides very fluid access to massive neuroimaging databases. It is extremely well suited for both the dissemination of massive 2D collections and direct exploratory analysis as part of a web service. Pivot has also been extremely helpful as part of quality control workflow and has enabled us to search for neuroanatomical differences and patterns of variation among strains of mice in ways that far surpass any other web interface.
References:
Lu L, Airey DC, Williams RW (2001) Complex trait analysis of the hippocampus: mapping and biometric analysis of two novel gene loci with specific effects on hippocampal structure in mice. J Neurosci 2001 21:3503-14
Rosen GD, Pung CJ, Owens CB, Caplow J, Kim H, Mozhui K, Lu L, Williams RW (2009) Genetic modulation of striatal volume by loci on Chrs 6 and 17 in BXD recombinant inbred mice. Genes Brain Behav 8:296-308
Yang RJ, Mozhui K, Karlsson RM, Cameron HA, Williams RW, Holmes A (2008) Variation in mouse basolateral amygdala volume is associated with differences in stress reactivity and fear learning. Neuropsychopharm 33:2595-2604
4 - MARYANN MARTONE
The INCF Program on Ontologies for Neural Structures
The goal of the program on Ontologies of Neural Structures is to promote data exchange and integration across disciplines, species, developmental stages, and structural scales by developing terminology standards and formal ontologies for neural structures. The current program comprises three task forces: the Structural Lexicon Task Force, the Neuronal Registry Task Force and the Representation and Deployment Task Force. These groups are working on formal definitions for brain regions and neurons through a formal definition of their properties. Demonstrations of the strategies and products will be debuted at the Kobe meeting.
5 - COLIN INGRAM
Practical metadata standards: Making data sharing and endurance feasible
Institute of Neuroscience, Newcastle University, UK
Neuroscience data are associated with a rich set of descriptive information that define the data structure, study design, and conditions of data acquisition (such as device characteristics, experimental protocol and parameters, behavioral paradigms, and subject/patient information) and for analyzed data, the statistical procedures applied. These metadata have a vital role in enabling an understanding of a dataset and without them any shared data have little value. To facilitate data archive, storage, sharing and re-use a number of areas of bioscience have developed agreed minimal metadata standards that have been adopted both by database curators and publishers. The INCF Program on Metadata Standards is examining the issues around developing metadata standards for neuroscience, including methods for efficient acquisition, domain-specific minimal standards, agreed terminology, formats and interoperability. The Oversight Committee has reviewed current schemas used to support neuroscience metadata and identified a number of the challenges associated with implementing an effective metadata structure. The major challenge facing the program is that, whilst it is broadly accepted that ‘minimal’ metadata are helpful for categorization or indexing of data, in most cases minimal metadata are insufficient to understand a study. On the other hand, describing every condition of an experiment in sufficient detail is difficult, and implementing detailed database schema can burden users and reduce the incentive to contribute, particularly for historical data. Thus, whilst one of the overall aims is to develop generic minimal requirements for reporting metadata, a key principle of the program is to develop approaches that will generate metadata of sufficient detail as to increase the value of the data both now and in the future. To this end we propose to develop metadata extraction tools that can automatically read metadata from acquisition devices or from existing file formats. This metadata extraction will also be applied to the analysis packages used for data processing in order that it is possible to understand the processes used to generate derived data. The program will also evaluate the opportunities for developing standards for the exchange of metadata and new methods for recording metadata at source (e.g. electronic lab books). Such automated extraction will reduce the overhead for generating the metadata whilst implementing a standardized acquisition method. A second principle underlying this program is that specific projects aimed at implementing metadata standards should be user-oriented, with a clear understanding of the value of the available data, the identity of the user community, and the potential outcomes arising from sharing. To this end the Oversight Committee has recommended establishing two Task Forces with a focus around the specific areas of fMRI and EEG where there are large user communities, employing similar methodology and where there can be considerable benefit from data sharing. The first Task Force will focus on the use case of developing a metadata schema to support resting fMRI data, and will develop methods for automating metadata acquisition at the level of the laboratory and ensuring interoperability between different data management systems. The second Task Force will focus on the use case of creating a domain standard for exchanging and describing EEG data, and will develop a framework for defining stimuli which are used in EEG experiments. Over the life time of the program these two use cases will be evaluated to determine the extent to which application of metadata standards can increase the access to shared data.
6 - DAVID VAN ESSEN
An informatics perspective on cerebral cortical connectivity and function
Washington University, St. Louis, USA
One of the great scientific challenges of the 21st century is to elucidate how the wiring of the human brain accounts for our unique cognitive capabilities and for individual differences in a wide variety of behavioral capacities. Recent advances on a variety of fronts, from data acquisition to neuroinformatics, will enable rapid progress on this daunting problem. The core data acquisition methods involve two powerful and complementary tools for analyzing human brain connectivity in vivo: tractography, which is based on diffusion imaging (DI), and functional connectivity analysis, which is based on spatial correlations in resting-state fMRI BOLD signals (R-fMRI). This presentation will discuss progress and challenges in using these methods to chart brain connectivity and to parcellate cerebral cortex based on differential patterns of connectivity (Fig. 1). This includes novel methods of data acquisition, analysis, and visualization, plus informatics enhancements for storing and communicating vast amounts of connectivity data. Several approaches to evaluating and validating connectivity data will be discussed. This includes comparisons with tracer-based connectivity maps acquired in nonhuman primates and registered to human cortex using plausible homologies between species. Collectively, these approaches are likely to yield deep insights into the fundamental basis of human brain function in health and disease.
7 - GARY EGAN
Neuroinformatics approaches for mapping striatal structures and cortico-striatal connections in the human brain
India Bohanna1,2, Nellie Georgiou-Karistianis3, Gary F. Egan 1,2
1 Florey Neuroscience Institute & 2 Centre for Neuroscience, University of Melbourne, Victoria 3010, Australia
3Experimental Neuropsychology Research Unit, School of Psychology and Psychiatry, Monash University, Clayton, Victoria, Australia
Major advances in our understanding of the human brain have resulted from neuroimaging research over the past two decades. The development of novel neuroinformatics approaches for the analysis of large neuroimaging datasets is enhancing our understanding of normal brain function as well as dysfunction in neurological and psychiatric diseases (1). Current models of cortico-striatal circuitry incorporate anatomically distinct parallel neural pathways projecting from the cerebral cortex to the striatum, each subserving a unique motor, cognitive or limbic function. Our current understanding of cortico-striatal functional neuroanatomy has been gained primarily from tract tracing studies in primates, but until recently these circuits have not been comprehensively demonstrated in the human brain (2).
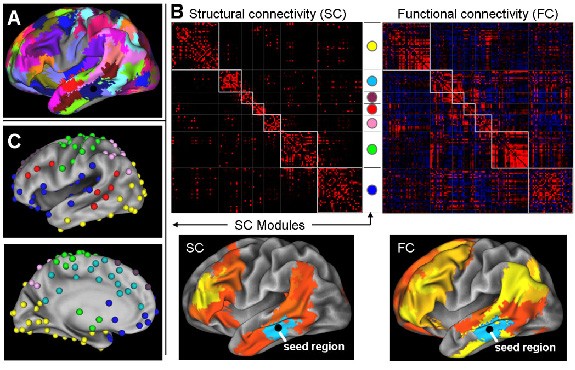
Diffusion Tensor Imaging (DTI) probabilistic tractography has been used to identify the spatial distribution of cortical connections within the human caudate and putamen. Cortical connections were topographically organized within the caudate and putamen, reflecting cerebral cortex organization, with significant spatial segregation between circuits. Both the caudate and putamen connected primarily with the prefrontal cortex highlighting a neuroanatomical difference between humans and primates in cortico-striatal circuitry, and suggests a possible role for the putamen in cognition.  Brain connectivity maps generated with advanced neuroinformatics techniques can facilitate detailed examination of the volume, connectivity and microstructure within individual striatal sub-regions and sub-circuits. Connectivity maps of the caudate and putamen are useful for examining the integrity of individual cortico-striatal circuits and/or striatal sub-regions, and can assist in investigating diseases characterised by regionally selective degeneration of the striatum, such as Huntington’s and Parkinson's diseases.
Brain connectivity maps generated with advanced neuroinformatics techniques can facilitate detailed examination of the volume, connectivity and microstructure within individual striatal sub-regions and sub-circuits. Connectivity maps of the caudate and putamen are useful for examining the integrity of individual cortico-striatal circuits and/or striatal sub-regions, and can assist in investigating diseases characterised by regionally selective degeneration of the striatum, such as Huntington’s and Parkinson's diseases.
Figure 1: (A) Three-dimensional segmentation of the human caudate (red) and putamen (blue). Organisation of cortical connections within the (B) caudate and (C) putamen. The cortical target regions of interest corresponding to each colour in (B & C) are shown at bottom. All segmentations are overlaid on the standard space MNI152 T1 weighted 1mm structural MR image. PF, Prefrontal; PM, Premotor; M1, Primary motor; S1; Somatosensory; PP, Posterior parietal; OCC, Occipital; TEM, Temporal
References
1. Bohanna I, et al. (2008) MRI identification of neuropathological biomarkers for HD. Brain Research Reviews 58:209-225
2. Alexander GE, et al. (1986). Parallel organization of functionally segregated circuits linking basal ganglia and cortex. Cited in Barton RA, Harvey PH (2000). Mosaic evolution of brain structure in mammals. Annu Rev Neurosci 9:357-381.
3. Draganski B, et al. (2008). Evidence for segregated and integrative connectivity patterns in the human Basal Ganglia. J Neurosci. 28:7143-52
4. Bohanna I, et al. (2010) Circuit-specific degeneration of the striatum in Huntington’s disease: a Diffusion Tensor Imaging study. Neuroimage in press (Proc. 16th Inter Conf Human Brain Mapping)
8 - SOO-YOUNG LEE
Perspetives on neuroinformatics as an important tool for an engineering goal - artificial cognitive system
Brain Science Research Center and Department of Electrical Engineering, KAIST, Daejeon, South Korea
We present two new research projects on neuroinformatics in Korea, i.e., Artificial Cognitive Systems (ACS) and Brain NeuroInformatics and Brain Engineering (BNIBE) projects. Both projects adopt the multidisciplinary integrative approach, and consist of brain signal measurements, computational models, and application test-beds of high-level cognitive functions.
The Artificial Cognitive Systems (ACS) will be based on Proactive Knowledge Development (PKD) and Self-Identity (SI) models. The PKD model provides bilateral interactions between robot and unknown environment (people, other robots, cyberspace, etc.). Based on the computational models of PKD and SI, we would like to build functional modules for Knowledge Represenation (Basic units of knowledge, i.e., features, and hierarchical network architecture based on the features), Knowledge Accumulation (Self-learning knowledge accumulation from environment), Situation Awareness (Recognition of unknown environment and situation based on knowledge, previous experience, and self-identity), Decision Making (Decision making based on situation, models of the users, and its own internal states), and Human Behavior (Action model for facial expression, hand motion, and speeches). The developed ACS will be tested against the new Turing Test for the situation awareness. The Test problems will consist of several video clips, and the performance of the ACSs will be compared against those of human with several levels of cognitive ability.
The Brain NeuroInformatics and Brain Engineering (BNIBE) project aims for computational models for cognitive functions and its applications to bilaterally-interactive man-machine interface (MMI). At the first phase of the BNIBE emphasizes the recognition of human intention, both explicit and implicit, from EEG, EOG, EMG, and audio-visual data. At the second phase, it will focus on the understanding of human behavior and its applications to real-time MMI.
9 - TAISHIN NOMURA
Integrated Bioscience with Dynamic Brain Platform
Graduate School of Engineering Science, Osaka University, Japan
The human genome sequencing was an epoch marking event in reductionist life science, liberating vast amount of experimental data. The challenge for the life sciences in the 21st century is to integrate this information into understanding of human physiology and pathology. In this context, advances in techniques for measurement of human body and functions including brain activities, information technology, and applied mathematics for modeling, simulating, and analyzing nonlinear dynamics continue their remarkable development. The integration of these fields and techniques is now moving the world towards a new generation of life science, where physiological and pathological information from the living human can be quantitatively described in silico across multiple scales of time and size and through diverse hierarchies of organization - from molecules to cells and organs to individuals. The physiome and systems biology in close interactions with bioinformatics and neuroinformatics represent such emerging biosciences. These new trends in biosciences share a common direction, namely “integrative” approach. This integrative approach is in stark contrast to linear and/or static approaches of the reductionist life science, allowing us to understand the mechanisms underlying physiological functions that will emerge through the dynamics of each element and large aggregations of the elements.
Integrative biosciences and engineering in its early stage aims at establishing frameworks and infrastructures for describing biological structure and physiological functions at multiple scales of time and space and then databasing and sharing them. To this end, standardized infrastructures that can deal with dynamics of proteins, cells, tissues, and organs, at multiple scales with multiple physics have been required. International communities such as for systems biology markup language (SBML), CellML, and Virtual Physiological Human (VPH) have promoted pioneering efforts to establish such frameworks. Through communication with these efforts, we have proposed insilicoML (ISML) possessing unique features that are compatible and complimentary to SBML and CellML. It is capable of representing hierarchical, modular, and morphological structure of physiological entities and functional operations of one module onto others. The integrated development environment, insilicoIDE (ISIDE), plays roles as a model composer, a browser, as well as a simulator of models written in ISML, CellML and SBML. The insilicoDB (ISDB) is a set of databases, including ISML-model DB, Morphology DB, and Time-series DB. They can be integrated on the ISIDE. The triplet (ISML, ISDB, and ISIDE) is available in the public domain as the physiome.jp open platform [1]. It can thus be enhancing the stream of model sharing and increasing diversity of building blocks useful for the integration. The physiome.jp has been collaborating with the Dynamic Brain Platform [2], a part of INCF J-Node, to exchange database contents of each platform and to provide standardized methodologies for constructing novel new models of brain functions by integrating models and data provided by the databases.
The development of the integrative biosciences and engineering will change conventional biology and medicine that have been based upon experience and expectation into predictive sciences. The predictive biosciences will have the capability to develop solutions based upon prior understanding of the dynamic mechanisms and the quantitative logic of physiology. Drug discovery, medical and welfare apparatus, and in silico trials for those will improve the development of products with higher efficiency, reliability and safety while reducing cost. They will also impact upon knowledge-intensive industries. It is thus essential for the future development of biosciences, clinical medicine, and industry, to establish infrastructures that are capable of playing core roles in the integration.
References
• http://www.physiome.jp
• http://dynamicbrain.neuroinf.jp/
RYUTARO HIMENO
Japan’s Next-Generation Supercomputer R&D Project and Grand Challenges in Life Science
Program for Computational Science, Integrated Simulation of Living Matter Group, RIKEN, Japan
MEXT and RIKEN started the Next-Generation Supercomputer R&D Project in 2006, of which goal is to develop a supercomputer with 10 Peta FLOPS in 2011FY. The supercomputer will be installed in a new supercomputer center in Kobe, where the buildings and facilities will be completed by the end of May, 2010. In this R&D project, not only hardware development but also software development in Nano Science and Life Science as Grand Challenge problems are planed and included. The Institute of Molecular Science leads the Nono Science Grand Challenge and RIKEN leads the Life Science one.
As the Life Science Grand Challenge, we have two approaches to achieve comprehensive understanding of life phenomena and to contribute to our society by creating medicine and developing medical/surgical treatment. Those approaches are 1) theoretical simulation approach and 2) data-driven approach. In addition to these approaches, we have a HPC team which is responsible to deliver maximum performance of the 10 Peta FLOPS supercomputer to tune the application software and to develop and provide tools for visualization and parallelization to other teams. These software tools are also available to anyone who will use the supercomputer. In 2007, we have added Brain and Neural Sytems Team lead by Prof. Ishii, Kyoto Universities.
Current status of supercomputer development and the application software development in Life Scinece will be shown in the talk.